What is Cre-Lox Recombination?
Over the past few decades, the science of Genomics has made leaps and bounds in terms of technological advancement. The development of gene editing technologies like the CRISPR Cas9 system and PCR allowed for the analyzing and the editing of the genome, which revolutionized fields like medicine, forensics, and even agriculture. One of these genome editing technologies, called Cre-Lox Recombination, is a site specific recombinase technology - a tool involving the use of recombinase enzymes to edit the genome, having the ability to carry out functions such as insertions, deletions, translocations, and inversions of target genes in the genome. Originally used by P1 bacteriophages in nature, Cre-Lox Recombination has been harnessed by scientists as a tool for genome manipulation and mainly used today in the field of neuroscience due to its ability to analyze complex cell types and piece together the functions of neural circuits.

What is Cre Recombinase?
Before explaining the mechanism of action of Cre - Lox recombination, it is important to first understand the components. As hinted before, the Cre-Lox system consists of two components: the recombination enzyme and the DNA. The recombination enzyme in this case is Cre Recombinase, or Cyclization Recombinase, a tyrosine recombinase enzyme with a topoisomerase-esque function. Like other members of the tyrosine recombinase family, Cre Recombinase possesses a tyrosine residue in its active site. Its job is to recognize sequences of DNA called Lox P sites and catalyze the process of site specific recombination. The enzyme’s ability to operate within a wide range of organisms and cellular environments add to its value as a tool for scientific research.
What is a LoxP site?
LoxP sites are also crucial components in the Cre-Lox recombination system. Standing for Locus of X over (cross over) in P1 bacteriophage, loxP sites are areas in DNA derived from P1 bacteriophage consisting of 34 base pairs, of which the first and last 13 base pairs are a palindromic sequence and are separated by a asymmetrical core sequence consisting of 8 base pairs. Because the loxP site consists of too many base pairs to appear naturally by chance, it possesses great specificity when it comes to genetic manipulation.
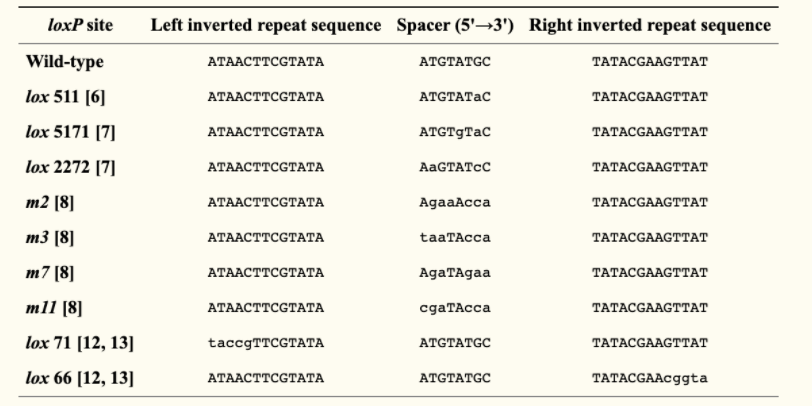
The loxP site’s core sequence helps determine the direction of the site, which will have an effect on the outcome of the cre lox recombination event. The usual loxP sequence, or the wild type, is ATAACTTCGTATA - ATGTAGCT - TATACGAAGTTAT, but the 8 bp sequence can be mutated to form different core sequences. There are also a variety of different genetically modified versions of loxP sites, which gives Cre Recombinase the ability to catalyze multiple loxP recombination events in a single system, as different loxP sequences are not cross compatible.
Mechanism of Action:
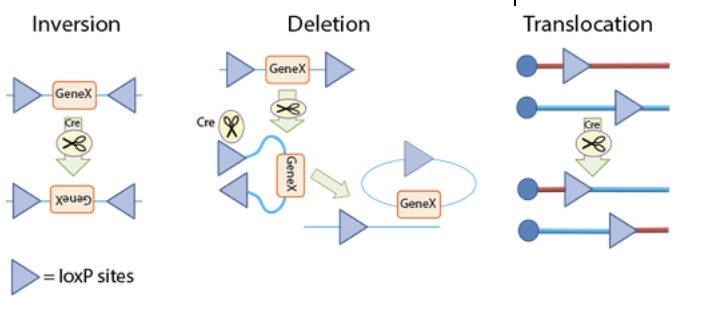
As mentioned before, this system uses an enzyme called Cre Recombinase to initiate site specific recombination between a pair of loxP sites. An important point to note is that these two loxP sites can be either located on the same strand or different strands of DNA. In Cre Recombination, there are two important conditions that affect the outcome: The position and the orientation of the loxP sites, but more on this later. In Cre-Lox Recombination, when an organism with loxP sites express Cre recombinase, the first step that happens is the recognition and binding of the Cre Recombinase on the first and last 13 base pairs on the LoxP site, which forms a dimer. The same event also happens to the second loxP site. Then, the strand or strands of DNA will reconfigure into a form that aligns the loxP sites into a parallel position and forms a tetrameter. The double stranded DNA is then broken by Cre Recombinase, rearranged, and reattached by DNA ligase. The end result of Cre-Lox Recombination activity is dependent on orientation and position of the loxP sites. If the two loxP sites are on the same strand of DNA and the orientation of the sites are of opposite directions, the gene between the two loxP sites will become inverted. If the loxP sites have the same orientation, the gene is cut out, and if the two loxP sites are on separate DNA strands, it is possible for translocation events to happen. LoxP sites on differing strands also can cause the merging of two plasmids in the presence of Cre Recombinase (as seen in the combination of two plasmids containing the lox 71 and and lox 66, image below). Another important note is that while the process of inverting the target gene is reversible, gene excision and translocation processes are not. There are two known factors playing roles in Cre-Lox Recombination efficiency. The first is the core sequence, where genetically engineered core sequences are usually not as high of recombination frequency as the wild type. The second factor that may influence the efficiency of action is the length of DNA between the loxP sites. The longer the length of DNA, the less efficient Cre-Lox Recombination becomes.
Significance and use:
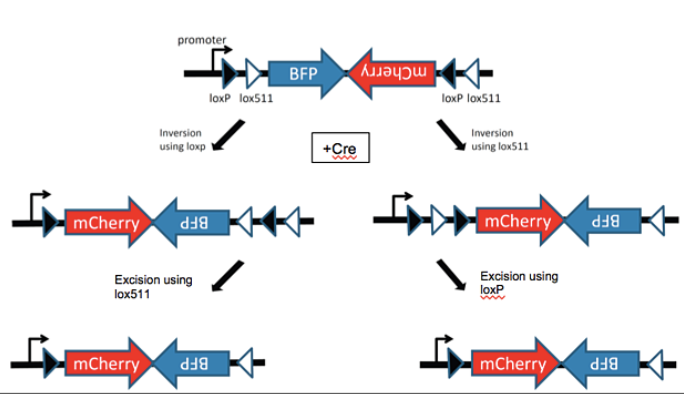
One of the most important and common uses of Cre-Lox recombination is Floxing, or when genes of interest are flanked by loxP. When genes are floxed, they are able to be expressed unless they are in the presence of Cre, at which point they would be disrupted or deleted. Floxing can help researchers identify phenotypes that are linked with specific genes. Another technique that can be used with Cre-Lox Recombination are FLEx switches, in which one of the loxP sites surrounding a floxed gene is floxed by a different loxP site variant. FLEx switches can invert a gene permanently by first inverting the sequences with the normal loxP sites surrounding the gene of interest then excising one of those loxP sites, which prevents further effects of Cre on the gene. Cre-Lox can also be used for limiting the production of certain cell products to certain cells at specific times. This is done through a process called Regulated Cre Expression wherein loxP sites are placed downstream of promoters active only in select cells and/or at select times, or by bringing Cre Recombinase to the cells at specific times. By regulating gene expression this way, scientists are able to identify gene activity in a much more controlled manner as well as being able to circumvent genetic embryonic lethalities.
Citations:
N. Sternberg, D. Hamilton, et al. “A Rapid in Vitro Method to Flip Back the Double-Floxed Inverted Open Reading Frame in a Plasmid.” BMC Biotechnology, BioMed Central, 1 Jan. 1981, bmcbiotechnol.biomedcentral.com/articles/10.1186/s12896-018-0462-x.
N;, Sauer B;Henderson. “Site-Specific DNA Recombination in Mammalian Cells by the Cre Recombinase of Bacteriophage P1.” Proceedings of the National Academy of Sciences of the United States of America, U.S. National Library of Medicine, pubmed.ncbi.nlm.nih.gov/2839833/.
N;, Sauer B;Henderson. “Site-Specific DNA Recombination in Mammalian Cells by the Cre Recombinase of Bacteriophage P1.” Proceedings of the National Academy of Sciences of the United States of America, U.S. National Library of Medicine, pubmed.ncbi.nlm.nih.gov/2839833/.